
Machine Learning Techniques to Analyze Dynamic Functional Neuroimaging Patterns Underlying Inhibitory Control Mechanisms
Analysis of functional Magnetic Resonance Images has allowed numerous insights in the way the brain functions. A large amount of data is being collected in numerous studies, however significant challenges remain towards automatic analysis of such data. These challenges include high levels of noise in the acquired signal, difficulties in registering scans between different subjects and different modalities, relatively small number of subjects per study and differences in protocol design and scanning equipment between studies. We hypothesize that unique patterns of variability in brain function can assist in identification of brain mechanisms rooted in neuroscientific prior knowledge. Such patterns will increase our understanding of brain connectivity and circuitry as we move iteratively between a-priori and exploratory means of describing functional brain circuits. We propose an integrated machine learning framework for the joint exploration of spatial, temporal and functional information for the analysis of fMRI signals, thus allowing the testing of hypotheses and development of applications that are not supported by traditional analysis methods. While our major focus is drug addiction and disorders of inhibitory control, we are testing our methods in datasets for other disorders such as major depression and autism.
In recent work with fMRI data we have demonstrated that it is possible to classify different groups of human subjects performing the same tasks based on the observed 3D fMRI BOLD images, through discovery of the appropriate features that capture the most discriminative differences in activation levels. We have validated our technique in multiple fMRI data sets. In prediction of behavior based on neuroimaging data, we search for the features which can unambiguously relate brain function with behavioral measurements. At the same time we have been exploring functional connectivity between brain regions by searching for conditional probabilistic dependencies between such regions, described by Gaussian Graphical Models, suitable to high-dimensional datasets. The method has guaranteed global minima, and does not require a-priori brain segmentation or selection of active regions. As a structure learning technique, the effect of confounding variables of brain region activations is removed, giving a clearer interpretation of possible dependencies between brain regions. Structure learning approaches will be useful in our quest to integrate spatiotemporally heterogeneous data sets such as MRI and PET. In immediate future work we plan to explore how domain knowledge and assumptions about common underlying neuropsychological processes can be used as constraints that allow us to combine similar (but not equal) features or dependencies across related datasets in multitask learning frameworks. Leveraging such related datasets will improve the stability of machine learning methods that are often encumbered by the difficulties posed by the high dimensionality in brain imaging datasets. In order to study larger cohorts of subjects we are also investigating resting state data for a number of disorders.
- Selected publications:
 Extracting brain regions from rest fMRI with Total-Variation constrained dictionary learning, Alexandre Abraham, Elvis Dohmatob, Bertrand Thirion, Dimitris Samaras, Gaël Varoquaux, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2013, Nagoya/Japan
Extracting brain regions from rest fMRI with Total-Variation constrained dictionary learning, Alexandre Abraham, Elvis Dohmatob, Bertrand Thirion, Dimitris Samaras, Gaël Varoquaux, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2013, Nagoya/Japan
 fMRI Analysis of Cocaine Addiction Using k-Support Sparsity, Gkirtzou K., Honorio J., Samaras D., Goldstein R., Blaschko M., In Proceedings of IEEE International Symposium on Biomedical Imaging 2013. California/USA.
fMRI Analysis of Cocaine Addiction Using k-Support Sparsity, Gkirtzou K., Honorio J., Samaras D., Goldstein R., Blaschko M., In Proceedings of IEEE International Symposium on Biomedical Imaging 2013. California/USA.
 Variable Selection for Gaussian Graphical Models, Honorio J., Samaras D., Rish I., Cecchi G., Artificial Intelligence and Statistics 2012. Canary Islands/Spain
Variable Selection for Gaussian Graphical Models, Honorio J., Samaras D., Rish I., Cecchi G., Artificial Intelligence and Statistics 2012. Canary Islands/Spain
[ BibTex] Can a Single Brain Region Predict a Disorder?, Honorio J., Tomasi D., Goldstein R., Leung H.C., Samaras D., IIEEE Transactions on Medical Imaging., 2012
Can a Single Brain Region Predict a Disorder?, Honorio J., Tomasi D., Goldstein R., Leung H.C., Samaras D., IIEEE Transactions on Medical Imaging., 2012
[ BibTex ] Multi-Task Learning of Gaussian Graphical Models, Honorio J., Samaras D., International Conference on Machine Learning, Haifa/Israel, 2010
Multi-Task Learning of Gaussian Graphical Models, Honorio J., Samaras D., International Conference on Machine Learning, Haifa/Israel, 2010
[ BibTex] Sparse and Locally Constant Gaussian Graphical Models, Honorio J., Ortiz L., Samaras D., Paragios N., Goldstein R.Panagopoulos A., Samaras D., Paragios N., Neural Information Processing Systems (NIPS'09), Vancouver/Canada, 2009
Sparse and Locally Constant Gaussian Graphical Models, Honorio J., Ortiz L., Samaras D., Paragios N., Goldstein R.Panagopoulos A., Samaras D., Paragios N., Neural Information Processing Systems (NIPS'09), Vancouver/Canada, 2009
[ BibTex] Task-Specific Functional Brain Geometry from Model Maps, Langs, G., Samaras, D., Paragios, N., Honorio, J., Alia-Klein, N., Tomasi, D., Volkow, N.D., Goldstein, R.Z, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) New York, NY, pp: 925-933, 2008
Task-Specific Functional Brain Geometry from Model Maps, Langs, G., Samaras, D., Paragios, N., Honorio, J., Alia-Klein, N., Tomasi, D., Volkow, N.D., Goldstein, R.Z, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) New York, NY, pp: 925-933, 2008
[ BibTex] Modeling Neuronal Interactivity using Dynamic Bayesian Networks, Lei Zhang, Dimitris Samaras, Nelly Alia-Klein, Nora Volkow, Rita Goldstein, Advances in Neural Information Processing (NIPS) 18, 2005, Vancouver, Canada, pp. 1593-1600.
Modeling Neuronal Interactivity using Dynamic Bayesian Networks, Lei Zhang, Dimitris Samaras, Nelly Alia-Klein, Nora Volkow, Rita Goldstein, Advances in Neural Information Processing (NIPS) 18, 2005, Vancouver, Canada, pp. 1593-1600. Machine Learning for Clinical Diagnosis from Functional Magnetic Resonance Imaging, Lei Zhang, Dimitris Samaras, Dardo Tomasi, Nora Volkow, Rita Goldstein, In IEEE Proc. of CVPR, pp. I: 1211-1217, 2005.
Machine Learning for Clinical Diagnosis from Functional Magnetic Resonance Imaging, Lei Zhang, Dimitris Samaras, Dardo Tomasi, Nora Volkow, Rita Goldstein, In IEEE Proc. of CVPR, pp. I: 1211-1217, 2005.
[ BibTex ] Exploiting Temporal Information in Functional Magnetic Resonance Imaging Brain Data, Lei Zhang, Dimitris Samaras, Nelly Klein, Dardo Tomasi, Lisa Cottone, Andreana Leskovjan, Nora Volkow, Rita Goldstein, In MICCAI, pp. 679-687, 2005.
Exploiting Temporal Information in Functional Magnetic Resonance Imaging Brain Data, Lei Zhang, Dimitris Samaras, Nelly Klein, Dardo Tomasi, Lisa Cottone, Andreana Leskovjan, Nora Volkow, Rita Goldstein, In MICCAI, pp. 679-687, 2005.
[ BibTex ]
- Collaborators: Rita Goldstein, Nelly Alia-Klein (Brookhaven National Lab)
- Students:
- Michail Misyrlis
- Former students:
- Jean Honorio
- Alexandros Panagopoulos
- Tejo Chalasani
- Lei Zhang
- Funding: NIH (NIDA)
- Keywords: Machine Learning, fMRI, Brain Imaging, Inhibitory Control
Extracting brain regions from rest fMRI with Total-Variation constrained dictionary learning
[publications]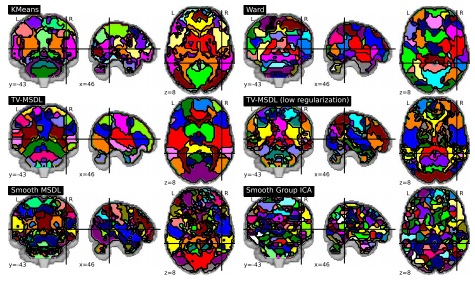
Spontaneous brain activity reveals mechanisms of brain function and dysfunction. Its population-level statistical analysis based on functional images often relies on the definition of brain regions that must summarize efficiently the covariance structure between the multiple brain networks. In this paper, we extend a network-discovery approach, namely dictionary learning, to readily extract brain regions. To do so, we introduce a new tool drawing from clustering and linear decomposition methods by carefully crafting a penalty. Our approach automatically extracts regions from rest fMRI that better explain the data and are more stable across subjects than reference decomposition or clustering methods.
Publications
 Extracting brain regions from rest fMRI with Total-Variation constrained dictionary learning, Alexandre Abraham, Elvis Dohmatob, Bertrand Thirion, Dimitris Samaras, Gaël Varoquaux, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2013, Nagoya/Japan
Extracting brain regions from rest fMRI with Total-Variation constrained dictionary learning, Alexandre Abraham, Elvis Dohmatob, Bertrand Thirion, Dimitris Samaras, Gaël Varoquaux, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2013, Nagoya/Japan
Functional Connectivity with Sparse Gaussian Graphical Models
[publications]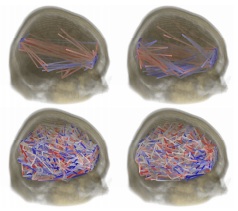
Structure learning techniques are very useful for analyzing complex datasets for which probabilistic dependencies are not known apriori. For instance, these techniques allow for modeling interactions between brain regions, based on measured activation levels through imaging. We propose the use of Gaussian graphical models for fully exploratory analysis applied to the voxel space, as it is suitable to high-dimensional datasets. The proposed structure learning algorithms are guaranteed to converge to the unique global minima, and do not require a-priori brain segmentation or selection of active regions. We present three regularizers for learning Gaussian graphical models from data: local constancy for datasets where variables correspond to a measurement in a manifold (silhouettes, motion trajectories, 2D and 3D images); variable selection for finding few interacting nodes from datasets with thousands of variables; and multi-task learning for a more efficient use of data which is available for multiple related tasks. For these regularizers, we show bounds of the eigenvalues of the optimal solution, convergence of block coordinate descent optimization, and connections to the continuous quadratic knapsack problem.
Publications
 Variable Selection for Gaussian Graphical Models, Honorio J., Samaras D., Rish I., Cecchi G., Artificial Intelligence and Statistics 2012. Canary Islands/Spain
Variable Selection for Gaussian Graphical Models, Honorio J., Samaras D., Rish I., Cecchi G., Artificial Intelligence and Statistics 2012. Canary Islands/Spain
[ BibTex] Multi-Task Learning of Gaussian Graphical Models, Honorio J., Samaras D., International Conference on Machine Learning, Haifa/Israel, 2010
Multi-Task Learning of Gaussian Graphical Models, Honorio J., Samaras D., International Conference on Machine Learning, Haifa/Israel, 2010
[ BibTex] Sparse and Locally Constant Gaussian Graphical Models, Honorio J., Ortiz L., Samaras D., Paragios N., Goldstein R.Panagopoulos A., Samaras D., Paragios N., Neural Information Processing Systems (NIPS'09), Vancouver/Canada, 2009
Sparse and Locally Constant Gaussian Graphical Models, Honorio J., Ortiz L., Samaras D., Paragios N., Goldstein R.Panagopoulos A., Samaras D., Paragios N., Neural Information Processing Systems (NIPS'09), Vancouver/Canada, 2009
[ BibTex]
fMRI Analysis of Cocaine Addiction Using k-Support Sparsity
[publications]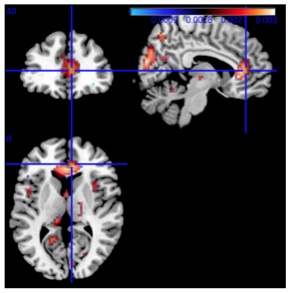
In this paper, we explore various sparse regularization tech- niques for analyzing fMRI data, such as LASSO, elastic net and the recently introduced k-support norm. Employing spar- sity regularization allow us to handle the curse of dimension- ality, a problem commonly found in fMRI analysis. We test these methods on real data of both healthy subjects as well as cocaine addicted ones and we show that although LASSO has good prediction, it lacks interpretability since the result- ing model is too sparse, and results are highly sensitive to the regularization parameter. We find that we can improve pre- diction performance over the LASSO using elastic net or the k-support norm, which is a convex relaxation to sparsity with an l2 penalty that is tighter than the elastic net. Elastic net and k-support norm overcome the problem of overly sparse solutions, resulting in both good prediction and interpretable solutions, while the k-support norm gave better prediction performance. Our experimental results support the general applicability of the k-support norm in fMRI analysis, both for prediction performance and interpretability.
Publications
 fMRI Analysis of Cocaine Addiction Using k-Support Sparsity, Gkirtzou K., Honorio J., Samaras D., Goldstein R., Blaschko M., In Proceedings of IEEE International Symposium on Biomedical Imaging 2013. California/USA.
fMRI Analysis of Cocaine Addiction Using k-Support Sparsity, Gkirtzou K., Honorio J., Samaras D., Goldstein R., Blaschko M., In Proceedings of IEEE International Symposium on Biomedical Imaging 2013. California/USA.
fMRI Analysis with Sparse Weisfeiler-Lehman Graph Statistics
[publications]fMRI analysis has most often been approached with linear methods. However, this disregards information encoded in the relation- ships between voxels. We propose to exploit the inherent spatial structure of the brain to improve the prediction performance of fMRI analysis. We do so in an exploratory fashion by representing the fMRI data by graphs. We use the Weisfeiler-Lehman algorithm to efficiently compute subtree features of the graphs. These features encode non-linear interactions be- tween voxels, which contain additional discriminative information that cannot be captured by a linear classifier. In order to make use of the effi- ciency of the Weisfeiler-Lehman algorithm, we introduce a novel pyramid quantization strategy to approximate continuously labeled graphs with a sequence of discretely labeled graphs. To control the capacity of the re- sulting prediction function, we utilize the elastic net sparsity regularizer. We validate our method on a cocaine addiction dataset showing a signif- icant improvement over elastic net and kernel ridge regression baselines and a reduction in classification error of over 14%. Source code is also available at https://gitorious.org/wlpyramid.
Publications
 fMRI Analysis with Sparse Weisfeiler-Lehman Graph Statistics, Medical Image Computing and Computer-Assisted Intervention, Workshop on Machine Learning in Medical Imaging 2013. Nagoya/Japan
fMRI Analysis with Sparse Weisfeiler-Lehman Graph Statistics, Medical Image Computing and Computer-Assisted Intervention, Workshop on Machine Learning in Medical Imaging 2013. Nagoya/Japan
Task-Specific Functional Brain Geometry from Model Maps
[publications]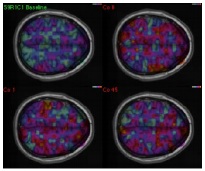
In this paper we propose model maps to derive and represent the intrinsic functional geometry of a brain from functional magnetic resonance imaging (fMRI) data for a specific task. Model maps represent the coherence of behavior of individual fMRI-measurements for a set of observations, or a time sequence. The maps establish a relation between individual positions in the brain by encoding the blood oxygen level dependent (BOLD) signal over a time period in a Markov chain. They represent this relation by mapping spatial positions to a new metric space, the model map. In this map the Euclidean distance between two points relates to the joint modeling behavior of their signals and thus the co-dependencies of the corresponding signals. The map reflects the functional as opposed to the anatomical geometry of the brain. It provides a quantitative tool to explore and study global and local patterns of resource allocation in the brain. To demonstrate the merit of this representation, we report quantitative experimental results on 29 fMRI time sequences, each with sub-sequences corresponding to 4 different conditions for two groups of individuals. We demonstrate that drug abusers exhibit lower differentiation in brain interactivity between baseline and reward related tasks, which could not be quantified until now.
Publications
 Task-Specific Functional Brain Geometry from Model Maps, Langs, G., Samaras, D., Paragios, N., Honorio, J., Alia-Klein, N., Tomasi, D., Volkow, N.D., Goldstein, R.Z, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) New York, NY, pp: 925-933, 2008
Task-Specific Functional Brain Geometry from Model Maps, Langs, G., Samaras, D., Paragios, N., Honorio, J., Alia-Klein, N., Tomasi, D., Volkow, N.D., Goldstein, R.Z, In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) New York, NY, pp: 925-933, 2008
[ BibTex]
Can a Single Brain Region Predict a Disorder?
[publications]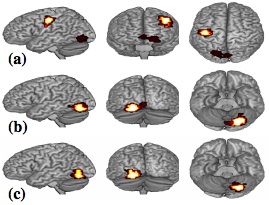
We perform prediction of diverse disorders (Cocaine Use, Schizophrenia and Alzheimer’s disease) in unseen subjects from brain fMRI. First, we show that for multi-subject prediction of simple cognitive states (e.g. motor vs. calculation and reading), voxels-as-features methods produce clusters that are similar for different leave-one-subject-out folds; while for group classification (e.g. cocaine addicted vs. control subjects), voxels are scattered and less stable. Therefore, we chose to use a single region per experimental condition and a majority vote classifier. Interestingly, our method outperforms state-of-the-art techniques. Our method can integrate multiple experimental conditions and successfully predict disorders in unseen subjects (leave-one-subjectout generalization accuracy: 89.3% and 90.9% for Cocaine Use, 96.4% for Schizophrenia and 81.5% for Alzheimer’s disease). Our experimental results not only span diverse disorders, but also different experimental designs (block design and event related tasks), facilities, magnetic fields (1.5Tesla, 3Tesla, 4Tesla) and speed of acquisition (interscan interval from 1600ms to 3500ms). We further argue that our method produces a meaningful low dimensional representation that retains discriminability.
Publications
 Can a Single Brain Region Predict a Disorder?, Honorio J., Tomasi D., Goldstein R., Leung H.C., Samaras D., IIEEE Transactions on Medical Imaging., 2012
Can a Single Brain Region Predict a Disorder?, Honorio J., Tomasi D., Goldstein R., Leung H.C., Samaras D., IIEEE Transactions on Medical Imaging., 2012
[ BibTex ] Simple Fully Automated Group Classification on Brain fMRI, Honorio J., Samaras D., Tomasi D., Goldstein R., IEEE International Symposium on Biomedical Imaging, Rotterdam/The Netherlands, 2010
Simple Fully Automated Group Classification on Brain fMRI, Honorio J., Samaras D., Tomasi D., Goldstein R., IEEE International Symposium on Biomedical Imaging, Rotterdam/The Netherlands, 2010
[ BibTex]
Modeling Neuronal Interactivity using Dynamic Bayesian Networks
[publications]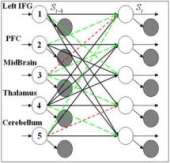
Functional Magnetic Resonance Imaging (fMRI) has enabled scientists to look into the active brain by providing sequences of 3D brain function images. However, interactivity between functional brain regions, is still little studied. In this paper, we contribute a novel framework for modeling the interactions between multiple active brain regions, using Dynamic Bayesian Networks (DBNs) as generative models for brain activation patterns. This framework is applied to modeling of neuronal circuits associated with reward. The novelty of our framework from a Machine Learning perspective lies in the use of DBNs to reveal the brain connectivity and interactivity. Such interactivity models which are derived from fMRI data are then validated through a classification task: separating drug addicted subjects from healthy non-drug-using controls. We employ and compare four different types of DBNs: Parallel Hidden Markov Models (PaHMM), Coupled Hidden Markov Models (CHMM), Fully-linked Hidden Markov Models (FHMM) and Dynamically Multi-Linked HMMs (DML-HMM). Moreover, we propose and compare two schemes of learning DML-HMMs. Experimental results show that by using DBNs, group classification can be performed even if the DBNs are constructed from as few as 5 brain regions. We also demonstrate that, by using the proposed learning algorithms, different DBN structures characterize drug addicted subjects vs. control subjects. This finding provides an independent test for the effect of psychopathology on brain function. In general, we demonstrate that incorporation of computer science principles into functional neuroimaging clinical studies provides a novel approach for probing human brain function.
Publications
 Modeling Neuronal Interactivity using Dynamic Bayesian Networks, Lei Zhang, Dimitris Samaras, Nelly Alia-Klein, Nora Volkow, Rita Goldstein, In NIPS, 2005.
Modeling Neuronal Interactivity using Dynamic Bayesian Networks, Lei Zhang, Dimitris Samaras, Nelly Alia-Klein, Nora Volkow, Rita Goldstein, In NIPS, 2005.
Machine Learning for Clinical Diagnosis from Functional Magnetic Resonance Imaging
[publications]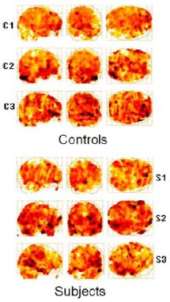
Functional Magnetic Resonance Imaging (fMRI) has enabled scientists to look into the active human brain. fMRI provides a sequence of 3D brain images with intensities representing brain activations. Standard techniques for fMRI analysis traditionally focused on finding the area of most significant brain activation for different sensations or activities. In this paper, we explore a new application of machine learning methods to a more challenging problem: classifying subjects into groups based on the observed 3D brain images when the subjects are performing the same task. Here we address the separation of drug-addicted subjects from healthy non-drug-using controls. In this paper, we explore a number of classification approaches. We introduce a novel algorithm that integrates side information into the use of boosting. Our algorithm clearly outperformed well established classifiers as documented in extensive experimental results. This is the first time that machine learning techniques based on 3D brain images are applied to a clinical diagnosis that currently is only performed through patient self-report. Our tools can therefore provide information not addressed by traditional analysis methods and substantially improve diagnosis.
Publications
 Machine Learning for Clinical Diagnosis from Functional Magnetic Resonance Imaging, Lei Zhang, Dimitris Samaras, Dardo Tomasi, Nora Volkow, Rita Goldstein, In IEEE Proc. of CVPR, pp. I: 1211-1217, 2005.
Machine Learning for Clinical Diagnosis from Functional Magnetic Resonance Imaging, Lei Zhang, Dimitris Samaras, Dardo Tomasi, Nora Volkow, Rita Goldstein, In IEEE Proc. of CVPR, pp. I: 1211-1217, 2005.
[ BibTex ]
Exploiting Temporal Information in Functional Magnetic Resonance Imaging Brain Data
[publications]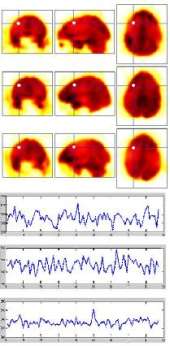
Functional Magnetic Resonance Imaging(fMRI) has enabled scientists to look into the active human brain, leading to a flood of new data, thus encouraging the development of new data analysis methods. In this paper, we contribute a comprehensive framework for spatial and temporal exploration of fMRI data, and apply it to a challenging case study: separating drug addicted subjects from healthy non-drug-using controls. To our knowledge, this is the first time that learning on fMRI data is performed explicitly on temporal information for classification. Experimental results demonstrate that, by selecting discriminative features, group classification can be successfully performed on our case study although training data are exceptionally high dimensional, sparse and noisy fMRI sequences. The classification performance can be significantly improved by incorporating temporal information into machine learning. Both statistical and neuroscientific validation of the method's generalization ability are provided. We demonstrate that incorporation of computer science principles into functional neuroimaging clinical studies, facilitates deduction about the behavioral probes from the brain activation data, thus providing a valid tool that incorporates objective brain imaging data into clinical classification of psychopathologies and identification of genetic vulnerabilities.
Publications
 Exploiting Temporal Information in Functional Magnetic Resonance Imaging Brain Data, Lei Zhang, Dimitris Samaras, Nelly Klein, Dardo Tomasi, Lisa Cottone, Andreana Leskovjan, Nora Volkow, Rita Goldstein, In MICCAI, pp. 679-687, 2005.
Exploiting Temporal Information in Functional Magnetic Resonance Imaging Brain Data, Lei Zhang, Dimitris Samaras, Nelly Klein, Dardo Tomasi, Lisa Cottone, Andreana Leskovjan, Nora Volkow, Rita Goldstein, In MICCAI, pp. 679-687, 2005.
[ BibTex ]
