Hi, this is Mousumi. I am a final year PhD student working with Prof. Fusheng Wang in the Computer Science Department at Stony Brook University, New York, USA. My primary research is focused on medical image analysis with machine learning and deep learning techniques. As a research intern, I got the opportunity to work at prestigious research institutions like IBM Research, DRDO. I am also the recipient of Graduate Assistance in Areas of National Need(GAANN) Fellowship Award.
I received my M.Tech. degree in Computer Science from University of Hyderabad(UoH) in India under the supervision of Prof. Arun Agarwal & Prof. Raghavendra Rao. I was a Gold Medalist for securing first rank in masters degree.
I am an active member of several women specific clubs at SBU like GWiSE, WPHD, SBCS among others and as GWiSE member, I mentor undergraduate students of WiSE group at SBU.
My primary research interest is artificial intelligence (AI) and big data analytics for healthcare, in particular, implementation of deep learning-based approaches for digital pathology imaging. My current research is based on the characterization of histopathological objects and exploring spatial relationship among a massive number of 2D and 3D objects from digital pathology images for clinically relevant correlations between the spatial patterns and disease progression through deep learning and computer vision. The goal is to create a novel framework to advance precision medicine through high resolution 3D computing pathology.
Publications
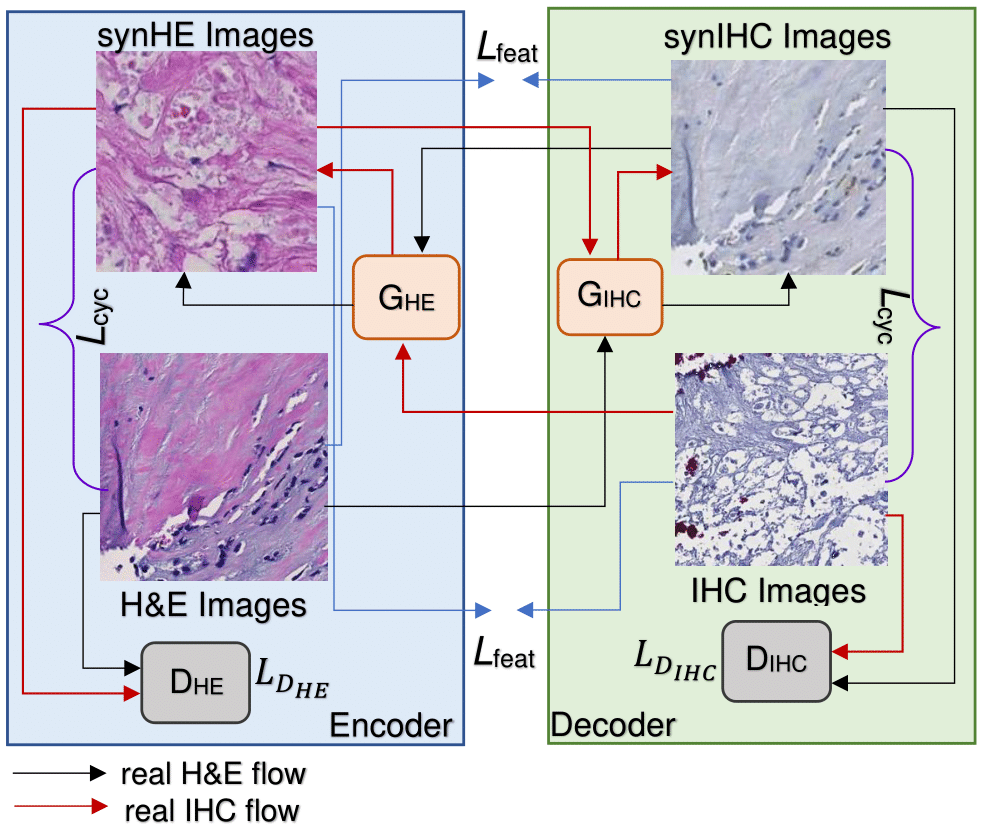
|
To register 2D serial sections from multiple stains (e.g., H\&E and IHC), we propose a novel translation based registration network CycGANRegNet using deep learning, which requires no prior deformation field information for training. We first generate synthetic IHC slides from H\&E slides through a robust image synthesis algorithm. The synthetic IHC images and the real IHC images are then registered through a Fully Convolutional Network with multi-scale deformable vector fields and a joint loss optimization for enhancing image alignment. We perform the registration at original image resolution with a patch-wide approach, thus tissue details at the highest resolution are retained in the results. CycGANRegNet outperforms both the state-of-the-art conventional and deep learning-based registration methods. |
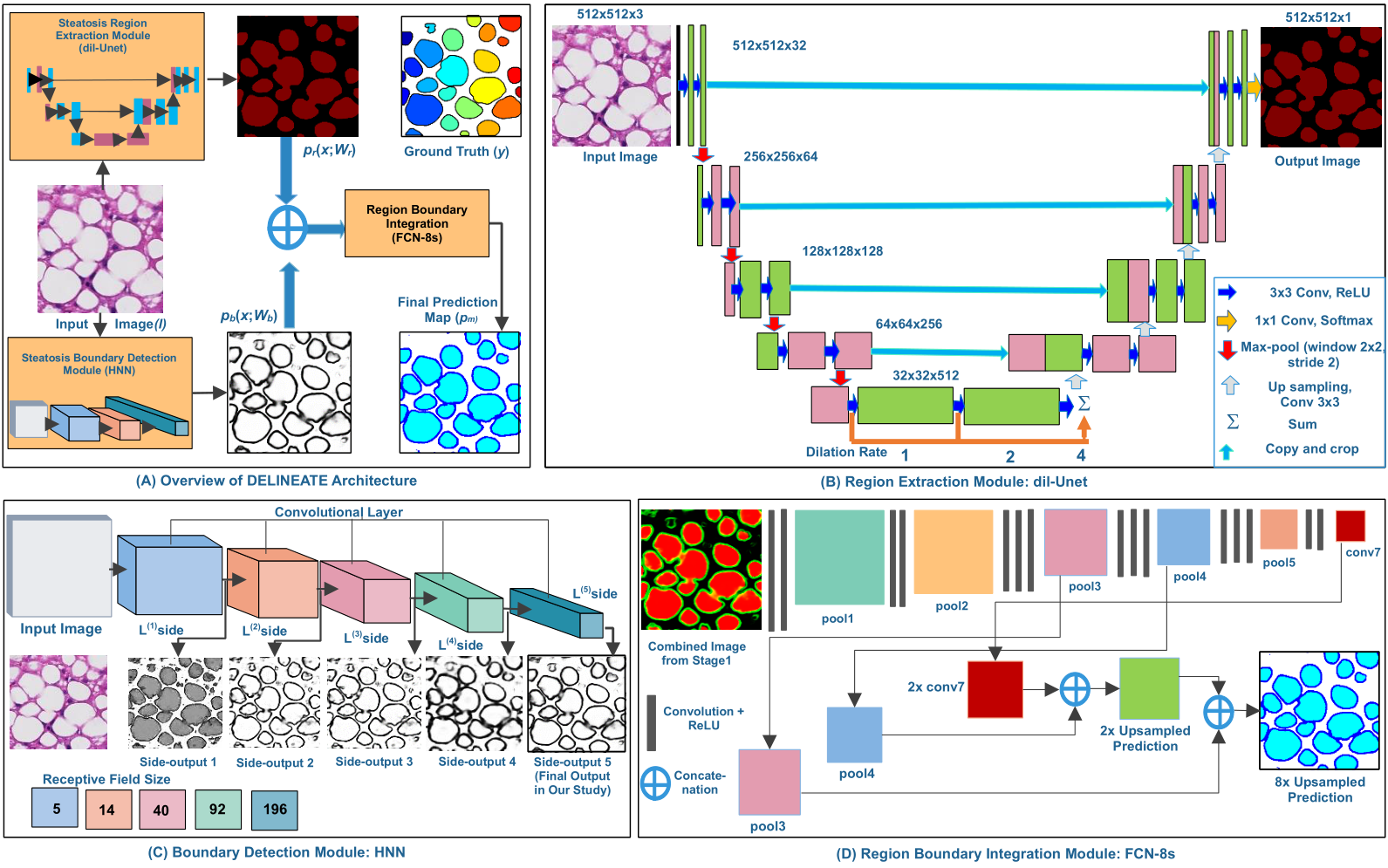
|
We propose a deep learning-based region-boundary integration network for overlapped steatosis droplets segmentation. The proposed network is designed using FCN, UNet and HNN. The strong correlation of our approach with other dimensions of data suggesting its potential to better support clinical decision through digital pathology. |
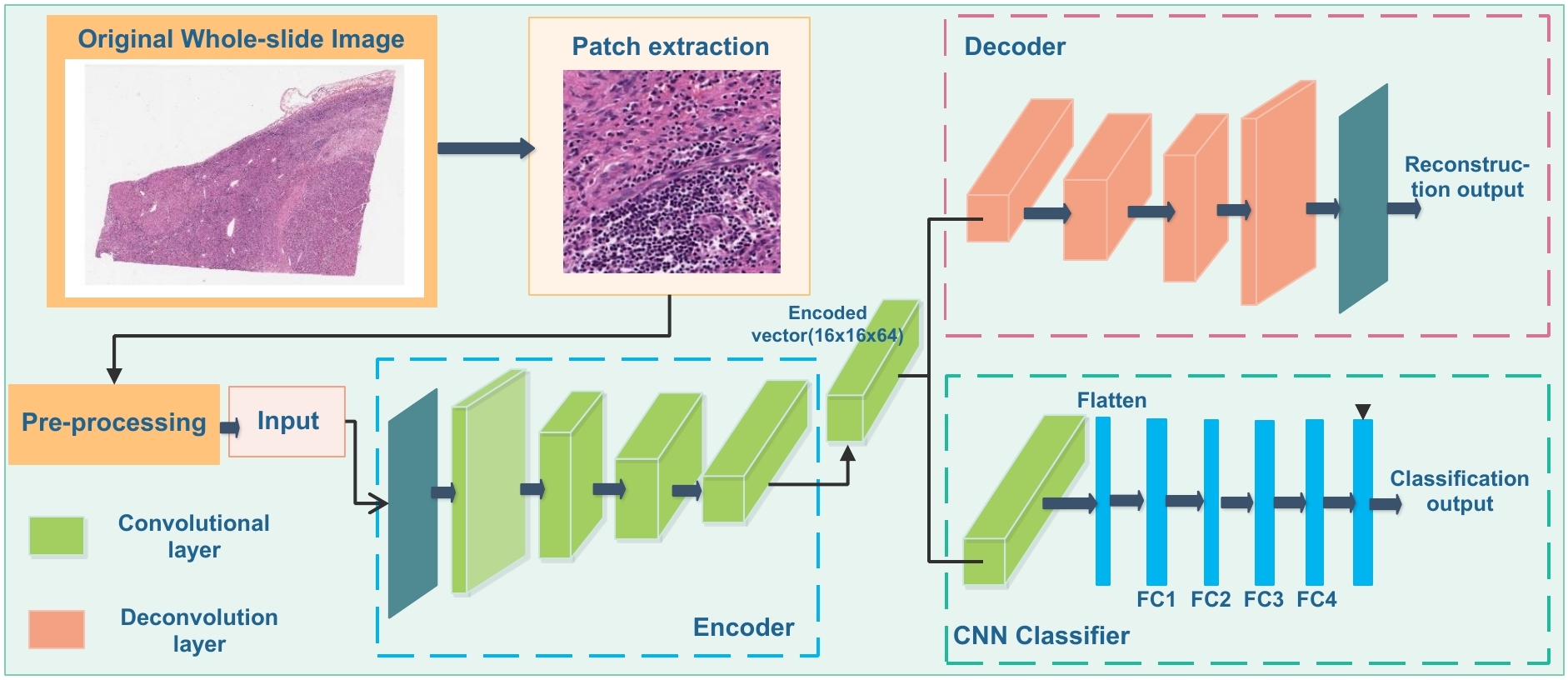
|
We present a multi-resolution convolutional autoencoder based model for viable tumor segmentation with a customized reconstruction loss for image reconstruction, followed by a classification module to classify each image patch. The resulting patch-based prediction results are spatially combined to generate the final segmentation results. Our proposed model presents superior performance to other benchmark models with extensive experiments. |
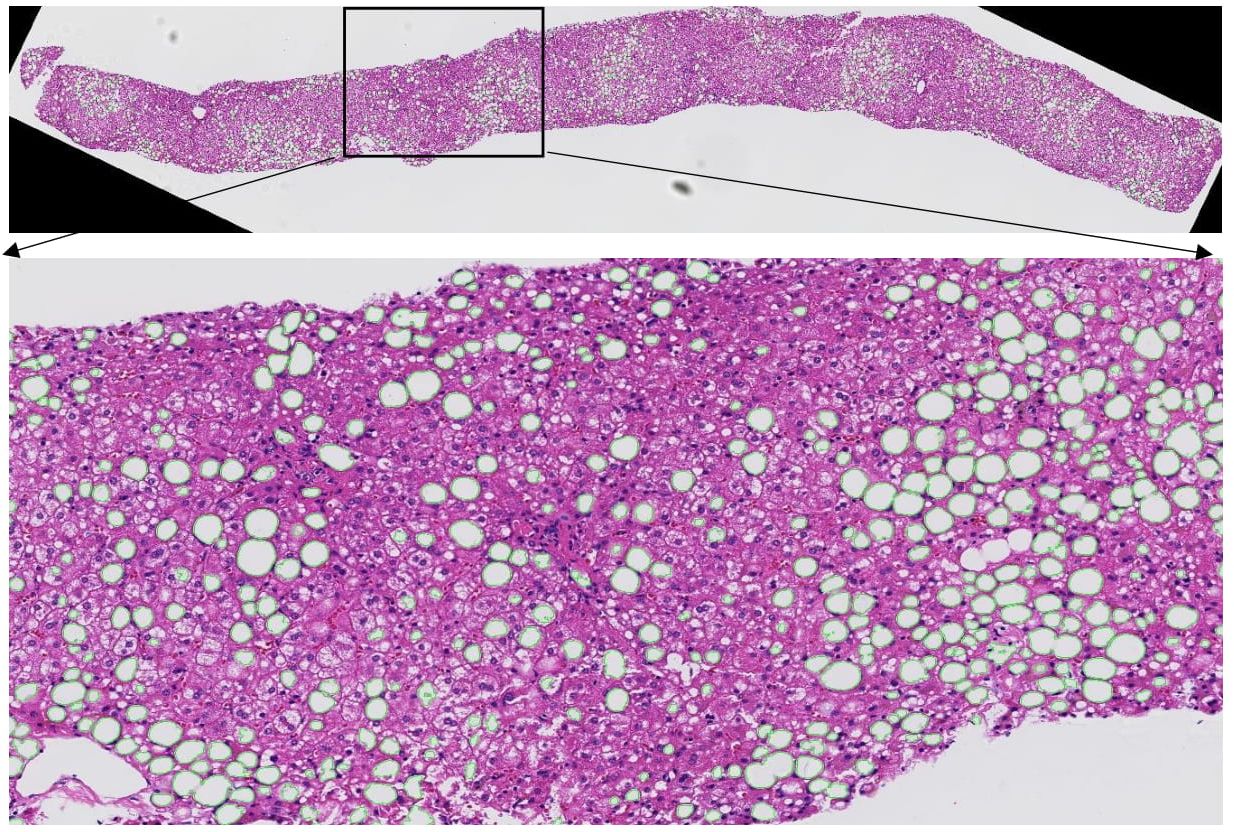
|
We developed a new image analysis method based on high curvature point detection and an ellipse fitting quality assessment method to segregate clumped micro-anatomic objects (steatosis) in whole-slide liver images. Our method demonstrates promising result for enhanced support of steatosis quantization during the pathology review for liver disease treatment. |
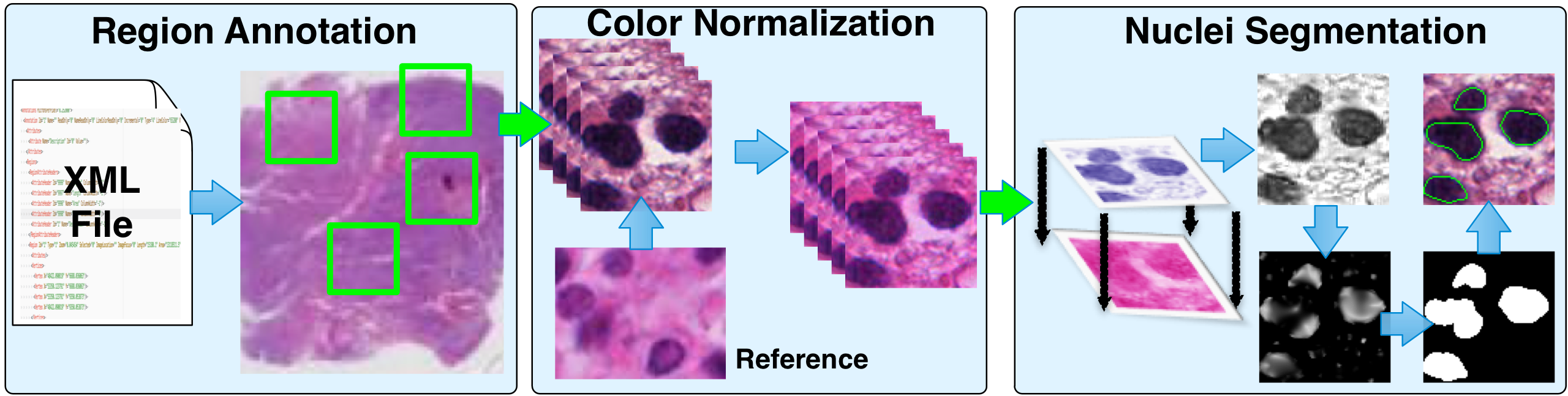
|
We propose a self-reliant and efficient analysis framework consisting of cell segmentation, feature computation, data pruning, dimensionality reduction and unsupervised clustering for quantitative analysis of cellular phenotypic difference across distinct molecular groups. Our analysis is generic and applicable to a wide set of cell-based bio-medical research. |
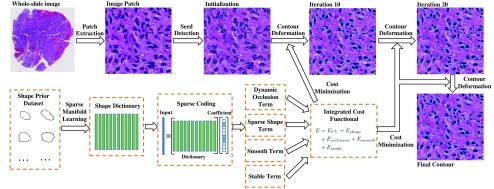
|
We propose a segmentation method for nuclei in glioblastoma histopathologic images based on a sparse shape prior guided variational level set framework. The proposed method is compared with other state-of-the-art methods and demonstrates good accuracy for nuclei detection and segmentation, suggesting its promise to support biomedical image-based investigations. |
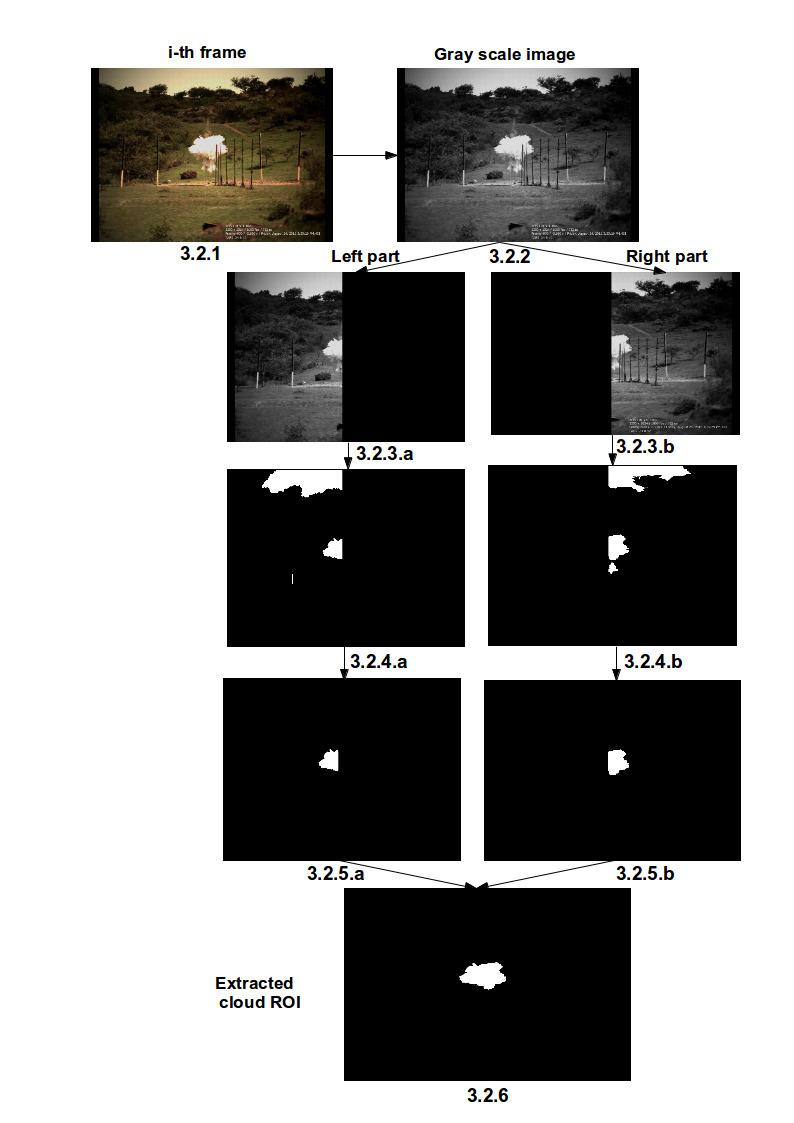
|
We demonstrated the understanding of cloud dynamics through image processing approach by analyzing video frames. The methodology involves selection of intensity band (HPF), cloud ROI extraction followed by cloud dimensions estimation and empirical model development to characterize the cloud dynamics. The developed model is found to be in good agreement with the reported model in literature. |
