BrainMiner:
ROI-Based Statistical Analysis, Representation, and
Discovery of Human Brain Functions
Development Team:
Faculty: Nora
Volkow (BNL Medical Group)
Klaus Mueller (Computer Science)
Wei Zhu (Applied Mathematics and Statistics)
Students: Tom Welsh and Jeffrey
Meade (CS)
Juan Li, Radha Panini, and Shurou (Sue) Wu (AMS)
Papers:
-
T. Welsh, K. Mueller, W. Zhu, N. Volkow, J. Meade, "Graphical
Strategies to Convey Functional Relationships in the Human Brain: A Case
Study", (to be presented) Visualization '01, San Diego, October 2001.
-
K. Mueller, T. Welsh, W. Zhu, J. Meade, N. Volkow, "BrainMiner:
A Visualization Tool for ROI-Based Discovery of Functional Relationships
in the Human Brain," New Paradigms in Information Visualization
and Manipulation (NPIVM) 2000, Washington DC, November, 2000.
View
this page in Romanian
Online Story:
Introduction
The overarching goal of this project is to investigate and discover correlational
relationships of different regions of the brain. Evidence for these relationships
are gathered by PET-imaging a number of human subjects, both under baseline
conditions and under the influence of a drug, in our case Ativan. The
correlations in the brain activity are calculated on the basis of predefined
anatomical regions-of-interest (ROIs), for now modeled as spherical regions.
The correlation coefficient is then employed to quantify similarity in response,
for various regions during an experimental setting. To account for inter-human
anatomical variability, each test subject's volumetric brain data is first transformed
into a common anatomical coordinate system (e.g, Talairach-Tournoux). Statistical
parameters that can be used to characterize various brain functions include:
-
The correlation (including the Pearson product-moment correlations, the
partial correlations and the canonical correlations) matrix..
-
The ROI clusters from the cluster analysis.
-
The principal components and the factor analysis output. (The PCA and the
FA are similar in their function and output, but different in their assumptions
and derivations).
-
Differential relationships such as the difference of two correlation matrices
(to view the change of functional relationships).
-
The times series.
The amount of statistical data can be enormous, and effective tools are
essential for the brain researcher to grasp and discover functional relationships
quickly from the statistical data. BrainMiner is a visualization
tool that facilitates this task.
Viewing in 2D
One approach to view statistical brain relationships is by overlaying the
statistically significant voxels on top of a high resolution MRI and view
the data as 2D slices, either in flip mode or side-by side:
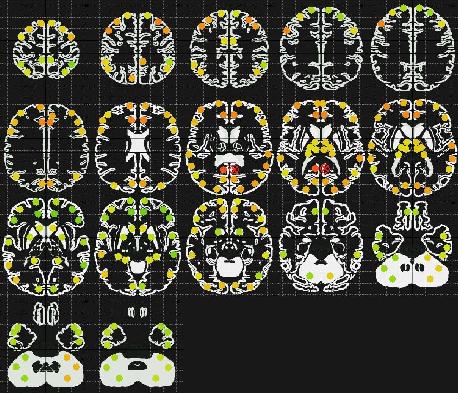
Fig. 1: The circular ROIs are colored according to their correlation
with respect to a root-ROI, marked by a red cross. The rainbow
color scheme is used, where the color blue stands for highly negative correlation
and the color red stands for highly positive correlation. Green and yellow
stand for mildly negative and positive correlations, respectively. There
are apparently no extremely strong correlations in this configuration.
The 2D approach works well as long as the axial dimension is not important.
However, the decomposition of the dataset into 2D slices for visualizing
3D relationships becomes limiting when relationships are widely spread
over the brain.
Viewing in 3D
To account for the problems with the 2D approach, we have also developed
(in addition to the 2D viewer) a 3D visualization interface that displays
correlational data for each ROI along with an MRI volume and a digitized
version of the Talairach atlas. Both can be sliced in 3 orthogonal directions
and can be overlaid on top of each other. Here is a screenshot of the Graphical
User Interface (GUI) of the system, where a basic view with a few ROIs
is shown:
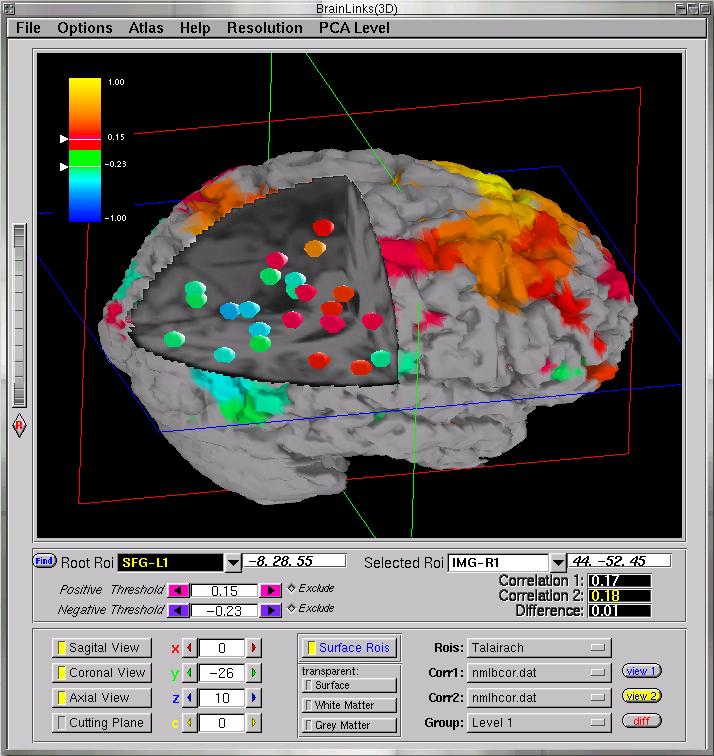
Fig. 2: Here we see the Graphical User Interface (GUI) of our
newly developed 3D brain visualization software, along with a basic view
of a small number of ROIs embedded into a cut-out area of a normalized/standardized
MRI brain. For now, all ROIs are spherical in shape. Similar to the 2D
viewer, the colors of the ROIs denote the strength of the correlational
relationship, on a rainbow scale. The root ROI is colored in yellow. The
GUI allows the user to slide the cutting planes up and down and back and
forth, to rotate the volume, and to select certain brain surfaces, such
as white matter, grey matter, and skull to be semi-transparently superimposed.
The correlation thresholds can also be selected, and many more features
are available.
The number of ROIs to be displayed, however, can become quite large (about
120-140), which poses challenging problems in the visualization task: In
a space too crowded with statistically significant ROIs, it becomes
very hard, if not impossible, for the user to tell the 3D positions of
the individual ROIs. To overcome these difficulties, a number of techniques
were investigated:
-
Superimposing a Talairach atlas slice that can be slid up and down the
volume:
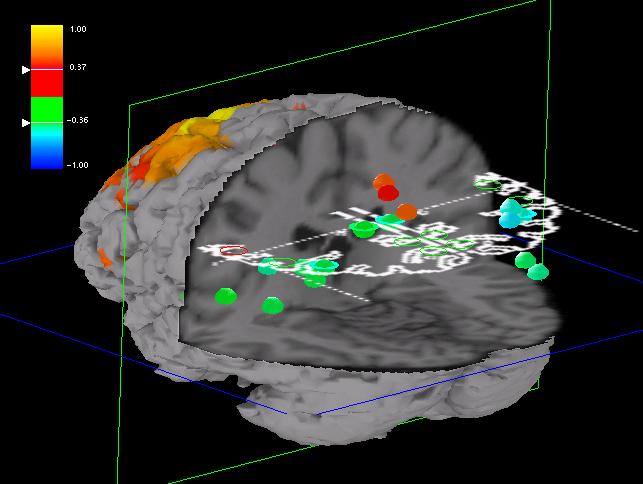
Fig 3: A movable sheet that shows the slice of the Talairach
atlas at the specified height. ROIs that intersect the sheet are highlighted
by a ring.
-
A single light source placed above the volume in a fixed position, providing
specular lighting cues for the height and depth of each ROI sphere (this
can be seen in Figs. 2 and 3).
-
Enhancing the ROIs by colored halos, where the colors code their
height and depth on a rainbow color scheme. The ROIs are connected by iso-lines
to the MRI volume cuts which suggests their position in 3D space:
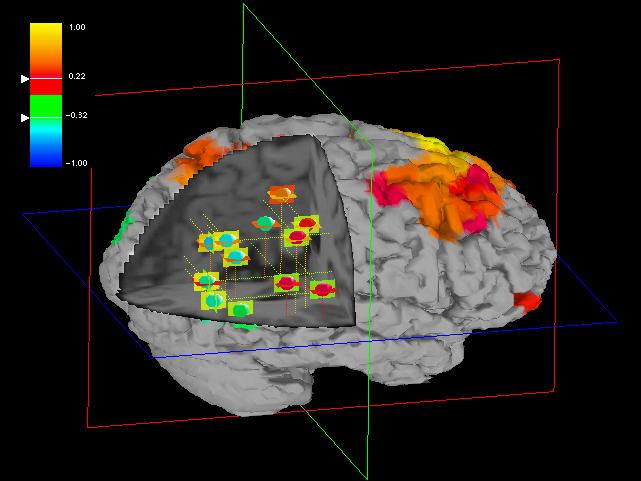
Fig. 4: ROI halos, painted in colors corresponding to
ROI height and depth (the rainbow color scheme is used). Dashed iso-height
and iso-depth lines emanate from the ROIs and pierce the MRI volume slices
at the ROI depth and height.
-
Projecting a colored grid onto the volume cuts, again encoding height and
depth on a rainbow map. Colored shadows cast onto the exposed volume slices
provide additional cues:
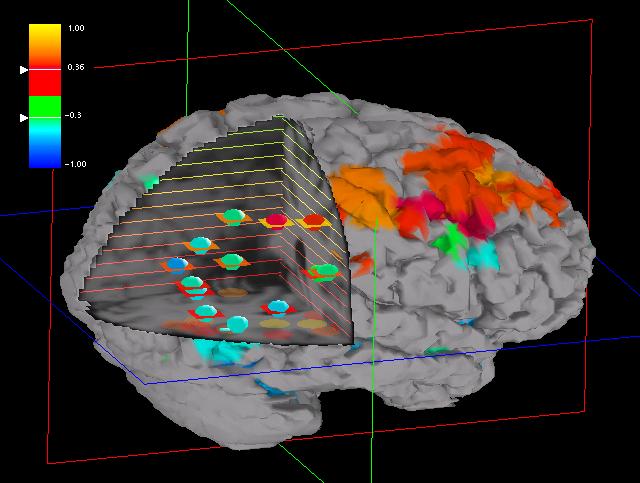
Fig. 5: Iso-lines, with height coded into rainbow colors, are
drawn onto two of the three orthogonal MRI volume cuts. The ROI halos are
coded in height using the same color scheme. The ROI position with respect
to the third MRI cut is suggested by shadows cast by the ROI spheres onto
that plane.
-
Projecting the ROIS onto the brain iso-surface, such as white or grey matter,
or skin:
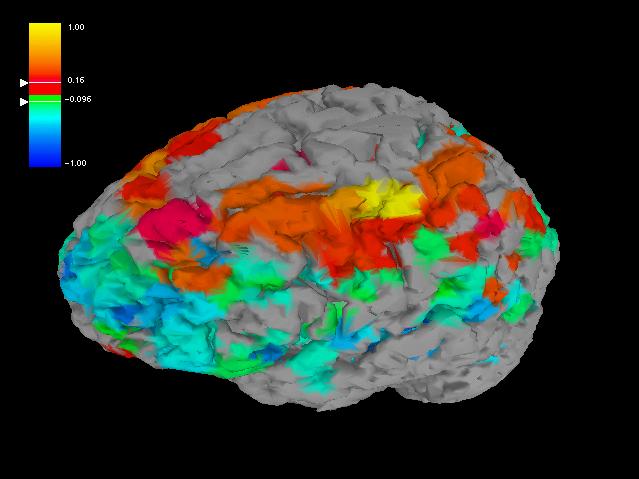
Fig. 6: Since the ROIs are mostly located close to the brain
surface, i.e. on the brain cortex, one can generate a comprehensive, EEG-like,
view by projecting the ROIs onto the cortex surface and paint the projection
in the correlation color.
-
Grouping ROI networks into composite polygonal objects, which reduces the
object complexity of the scene:
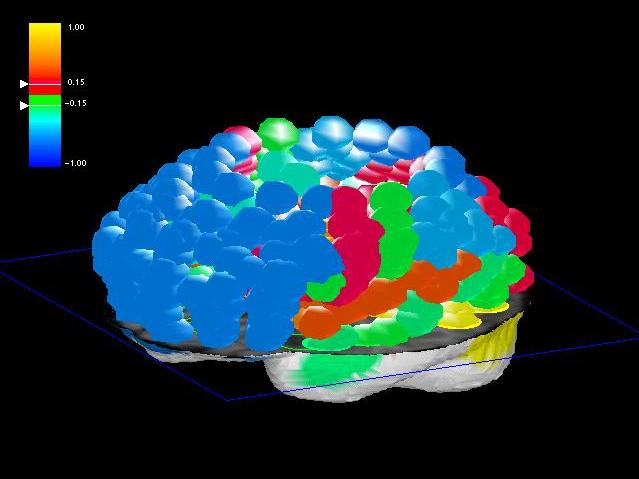
Fig. 7: For now we simply increased the radius of the ROI spheres
until they just touched. This approach is rather effective. For the future
we plan to estimate the actual hull of a set of ROIs of similar brain function
and display this hull as a polymesh.
Results
To test the software and develop its capabilities, PET fluoro-18-deoxyglucose
(FDG) images were analyzed and displayed for two major drug addiction studies.
The first study included 30 subjects the second included 40 subjects both
under baseline and drug conditions. Metabolic activity was measured for
each subject as the average intensity signal for a given ROI defined manually
by a trained medical doctor. ROI locations for the first study included
424 anatomically significant regions while the second study included 120
regions. Correlation matrices were generated for each of these ROI datasets.
Three major statistical measures were subsequently generated given the
correlation matrix for each study: 1) Principle Component Analysis, 2)
ROI cluster analysis and 3) Factor Analysis. Visualization software was
compiled using libraries for OpenGL, the Fast Light Toolkit (FLTK) and
written in C++. Versions were compiled for Irix 6.4, Windows NT 4.0 and
Linux and run on an SGI O2/R1000, a Pentium II class laptop and a Pentium
233 respectively. The visualization interface displayed a 3D MRI volume
representative of Talairach coordinates which was sliced as three half
planes of axial, coronal and saggital views. The slices were drawn using
2D texture mapping. The user could define the slice location along these
three axis by dragging each slice with a mouse button. The surface of the
brain was also displayed as a polygonal mesh generated from the Marching
Cubes algorithm. ROIs were drawn as spheres but were obscured by the surface
depending on the current slice locations. Correlation value was represented
for each ROI as a color intensity in relation to a selected "root" ROI.
A side window also displayed the current slice as either the Talairach
atlas digitized or the MRI volume along with a 2D representation of the
ROIs (circles). In addition, all objects could be rotated together with
the mouse (trackball interface) to provide a viewpoint from any direction.
Conclusions
BrainMiner
allows interactive exploration of both the 120x120 and 424x424 correlational
data on many platforms. It is cuurently in use by a number of brain researchers
and is being refined almost on an hourly basis.
This page has been translated into Norwegian
by globe-views.com
Back to research page







